diff --git a/MEDimage/__init__.py b/MEDimage/__init__.py
index 5db0645..647477d 100644
--- a/MEDimage/__init__.py
+++ b/MEDimage/__init__.py
@@ -14,7 +14,7 @@
logging.getLogger(__name__).addHandler(stream_handler)
__author__ = "MEDomicsLab consortium"
-__version__ = "0.9.4"
+__version__ = "0.9.7"
__copyright__ = "Copyright (C) MEDomicsLab consortium"
__license__ = "GNU General Public License 3.0"
__maintainer__ = "MAHDI AIT LHAJ LOUTFI"
diff --git a/MEDimage/filters/TexturalFilter.py b/MEDimage/filters/TexturalFilter.py
index 0b8c0d6..c98293a 100644
--- a/MEDimage/filters/TexturalFilter.py
+++ b/MEDimage/filters/TexturalFilter.py
@@ -7,8 +7,8 @@
import pycuda.driver as cuda
from pycuda.autoinit import context
from pycuda.compiler import SourceModule
-except ImportError:
- print("PyCUDA is not installed. Please install it to use the textural filters.")
+except Exception as e:
+ print("PyCUDA is not installed. Please install it to use the textural filters.", e)
import_failed = True
from ..processing.discretisation import discretize
diff --git a/MEDimage/learning/RadiomicsLearner.py b/MEDimage/learning/RadiomicsLearner.py
index 0dbc063..b112a4d 100644
--- a/MEDimage/learning/RadiomicsLearner.py
+++ b/MEDimage/learning/RadiomicsLearner.py
@@ -295,7 +295,7 @@ def train_xgboost_model(
optimal_threshold: float = None,
optimization_metric: str = 'MCC',
method : str = "pycaret",
- use_gpu: bool = True,
+ use_gpu: bool = False,
seed: int = None,
) -> Dict:
"""
@@ -338,6 +338,7 @@ def train_xgboost_model(
fold=5,
target=temp_data.columns[-1],
use_gpu=use_gpu,
+ feature_selection_estimator="xgboost",
session_id=seed
)
diff --git a/MEDimage/learning/Results.py b/MEDimage/learning/Results.py
index 3c165df..2c69654 100644
--- a/MEDimage/learning/Results.py
+++ b/MEDimage/learning/Results.py
@@ -417,9 +417,7 @@ def get_model_performance(
def get_optimal_level(
self,
path_experiments: Path,
- experiment: str,
- modalities: List,
- levels: List,
+ experiments_labels: List[str],
metric: str = 'AUC_mean',
p_value_test: str = 'wilcoxon',
aggregate: bool = False,
@@ -429,10 +427,8 @@ def get_optimal_level(
Args:
path_experiments (Path): Path to the folder containing the experiments.
- experiment (str): Name of the experiment to plot. Will be used to find the results.
- modalities (List): List of imaging modalities to include in the plot.
- levels (List): List of radiomics levels to include in plot. For example: ['morph', 'intensity'].
- You can also use list of variants to plot the best variant for each level. For example: [['morph', 'morph5'], 'intensity'].
+ experiments_labels (List): List of experiments labels to use for the plot. including variants is possible. For
+ example: ['experiment1_morph_CT', ['experiment1_intensity5_CT', 'experiment1_intensity10_CT'], 'experiment1_texture_CT'].
metric (str, optional): Metric to plot. Defaults to 'AUC_mean'.
p_value_test (str, optional): Method to use to calculate the p-value. Defaults to 'wilcoxon'.
Available options:
@@ -449,15 +445,47 @@ def get_optimal_level(
"""
assert metric.split('_')[0] in list_metrics, f'Given metric {list_metrics} is not in the list of metrics. Please choose from {list_metrics}'
- # Initialization
+ # Extract modalities and initialize the dictionary
+ if type(experiments_labels[0]) == str:
+ experiment = '_'.join(experiments_labels[0].split('_')[:-2])
+ elif type(experiments_labels[0]) == list:
+ experiment = '_'.join(experiments_labels[0][0].split('_')[:-2])
+
+ modalities = set()
+ for exp_label in experiments_labels:
+ if isinstance(exp_label, str):
+ modalities.add(exp_label.split("_")[-1])
+ elif isinstance(exp_label, list):
+ for sub_exp_label in exp_label:
+ modalities.add(sub_exp_label.split("_")[-1])
+ else:
+ raise ValueError(f'experiments_labels must be a list of strings or a list of list of strings, given: {type(exp_label)}')
+
+ levels_dict = {modality: [] for modality in modalities}
optimal_lvls = [""] * len(modalities)
+ # Populate the dictionary
+ variants = []
+ for label in experiments_labels:
+ if isinstance(label, str):
+ modality = label.split("_")[-1]
+ levels_dict[modality].append(label.split("_")[-2])
+ elif isinstance(label, list):
+ modality = label[0].split("_")[-1]
+ variants = []
+ for sub_label in label:
+ variants.append(sub_label.split("_")[-2])
+ levels_dict[modality] += [variants]
+
# Prepare the data for the heatmap
for idx_m, modality in enumerate(modalities):
best_levels = []
results_dict_best = dict()
results_dicts = []
best_exp = ""
+ levels = levels_dict[modality]
+
+ # Loop over the levels and find the best variant for each level
for level in levels:
metric_compare = -1.0
if type(level) != list:
@@ -496,7 +524,7 @@ def get_optimal_level(
# Statistical analysis
# Initializations
- optimal_lvls[idx_m] = best_levels[0]
+ optimal_lvls[idx_m] = experiment + "_" + best_levels[0] + "_" + modality
init_metric = heatmap_data[0][0]
idx_d = 0
start_level = 0
@@ -523,7 +551,7 @@ def get_optimal_level(
# If p-value is less than 0.05, change starting level
if p_value <= 0.05:
- optimal_lvls[idx_m] = best_levels[idx_d+1]
+ optimal_lvls[idx_m] = experiment + "_" + best_levels[idx_d+1] + "_" + modality
init_metric = metric_val
start_level = idx_d + 1
@@ -616,9 +644,7 @@ def plot_features_importance_histogram(
def plot_heatmap(
self,
path_experiments: Path,
- experiment: str,
- modalities: List,
- levels: List,
+ experiments_labels: List[str],
metric: str = 'AUC_mean',
stat_extra: list = [],
plot_p_values: bool = True,
@@ -633,10 +659,8 @@ def plot_heatmap(
Args:
path_experiments (Path): Path to the folder containing the experiments.
- experiment (str): Name of the experiment to plot. Will be used to find the results.
- modalities (List): List of imaging modalities to include in the plot.
- levels (List): List of radiomics levels to include in plot. For example: ['morph', 'intensity'].
- You can also use list of variants to plot the best variant for each level. For example: [['morph', 'morph5'], 'intensity'].
+ experiments_labels (List): List of experiments labels to use for the plot. including variants is possible. For
+ example: ['experiment1_morph_CT', ['experiment1_intensity5_CT', 'experiment1_intensity10_CT'], 'experiment1_texture_CT'].
metric (str, optional): Metric to plot. Defaults to 'AUC_mean'.
stat_extra (list, optional): List of extra statistics to include in the plot. Defaults to [].
plot_p_values (bool, optional): If True plots the p-value of the choosen test. Defaults to True.
@@ -657,9 +681,42 @@ def plot_heatmap(
None.
"""
assert metric.split('_')[0] in list_metrics, f'Given metric {list_metrics} is not in the list of metrics. Please choose from {list_metrics}'
+
+ # Extract modalities and initialize the dictionary
+ if type(experiments_labels[0]) == str:
+ experiment = '_'.join(experiments_labels[0].split('_')[:-2])
+ elif type(experiments_labels[0]) == list:
+ experiment = '_'.join(experiments_labels[0][0].split('_')[:-2])
+
+ modalities = set()
+ for exp_label in experiments_labels:
+ if isinstance(exp_label, str):
+ modalities.add(exp_label.split("_")[-1])
+ elif isinstance(exp_label, list):
+ for sub_exp_label in exp_label:
+ modalities.add(sub_exp_label.split("_")[-1])
+ else:
+ raise ValueError(f'experiments_labels must be a list of strings or a list of list of strings, given: {type(exp_label)}')
+
+ levels_dict = {modality: [] for modality in modalities}
+
+ # Populate the dictionary
+ variants = []
+ for label in experiments_labels:
+ if isinstance(label, str):
+ modality = label.split("_")[-1]
+ levels_dict[modality].append(label.split("_")[-2])
+ elif isinstance(label, list):
+ modality = label[0].split("_")[-1]
+ variants = []
+ for sub_label in label:
+ variants.append(sub_label.split("_")[-2])
+ levels_dict[modality] += [variants]
# Prepare the data for the heatmap
fig, axs = plt.subplots(len(modalities), figsize=figsize)
+
+ # Heatmap conception
for idx_m, modality in enumerate(modalities):
# Initializations
best_levels = []
@@ -667,6 +724,7 @@ def plot_heatmap(
results_dicts = []
best_exp = ""
patients_count = dict.fromkeys([modality])
+ levels = levels_dict[modality]
# Loop over the levels and find the best variant for each level
for level in levels:
@@ -1502,7 +1560,7 @@ def plot_original_level_tree(
fig.tight_layout()
# Save the plot (Mandatory, since the plot is not well displayed on matplotlib)
- fig.savefig(path_experiments / f'Original_level_{experiment}_{level}_{modality}_explanation.png', dpi=300)
+ fig.savefig(path_experiments / f'Original_level_{experiment}_{level}_{modality}_explanation_tree.png', dpi=300)
def plot_lf_level_tree(
self,
diff --git a/MEDimage/learning/Stats.py b/MEDimage/learning/Stats.py
index cf2e8d5..8af2a54 100644
--- a/MEDimage/learning/Stats.py
+++ b/MEDimage/learning/Stats.py
@@ -32,7 +32,7 @@ class Stats:
levels (List): List of radiomics levels to analyze.
modalities (List): List of modalities to analyze.
"""
- def __init__(self, path_experiment: Path, experiment: str, levels: List, modalities: List):
+ def __init__(self, path_experiment: Path, experiment: str = "", levels: List = [], modalities: List = []):
# Initialization
self.path_experiment = path_experiment
self.experiment = experiment
diff --git a/MEDimage/learning/ml_utils.py b/MEDimage/learning/ml_utils.py
index 0b496c0..edaa744 100644
--- a/MEDimage/learning/ml_utils.py
+++ b/MEDimage/learning/ml_utils.py
@@ -466,8 +466,8 @@ def cross_validation_split(
train_data_list.append(patient_ids[train_indices])
test_data_list.append(patient_ids[test_indices])
- train_data_array = np.array(train_data_list)
- test_data_array = np.array(test_data_list)
+ train_data_array = np.array(train_data_list, dtype=object)
+ test_data_array = np.array(test_data_list, dtype=object)
return train_data_array, test_data_array
diff --git a/MEDimage/wrangling/DataManager.py b/MEDimage/wrangling/DataManager.py
index 276ecec..348bb7e 100644
--- a/MEDimage/wrangling/DataManager.py
+++ b/MEDimage/wrangling/DataManager.py
@@ -652,11 +652,12 @@ def update_from_csv(self, path_csv: Union[str, Path] = None) -> None:
self.csv_data = csv_data
self.summarize()
- def summarize(self):
+ def summarize(self, retrun_summary: bool = False) -> None:
"""Creates and shows a summary of processed scans organized by study, institution, scan type and roi type
Args:
- None
+ retrun_summary (bool, optional): If True, will return the summary as a dictionary.
+
Returns:
None
"""
@@ -728,6 +729,9 @@ def count_scans(summary):
}, ignore_index=True)
print(summary_df.to_markdown(index=False))
+ if retrun_summary:
+ return summary_df
+
def __pre_radiomics_checks_dimensions(
self,
path_data: Union[Path, str] = None,
diff --git a/README.md b/README.md
index 327c891..8a1bbee 100644
--- a/README.md
+++ b/README.md
@@ -3,11 +3,11 @@
 [](https://www.python.org/downloads/release/python-380/)
-[](https://test.pypi.org/project/medimage-pkg/0.2.0/)
-[](https://github.com/MEDomics-UdeS/MEDimage/actions/workflows/python-app.yml)
+[](https://pypi.org/project/medimage-pkg/)
+[](https://github.com/MahdiAll99/MEDimage/actions/workflows/python-app.yml)
[](https://medimage.readthedocs.io/en/latest/?badge=latest)
[](LICENSE)
-[](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/tutorial/DataManager-Tutorial.ipynb)
+[](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/tutorial/DataManager-Tutorial.ipynb)
@@ -27,13 +27,13 @@
## 1. Introduction
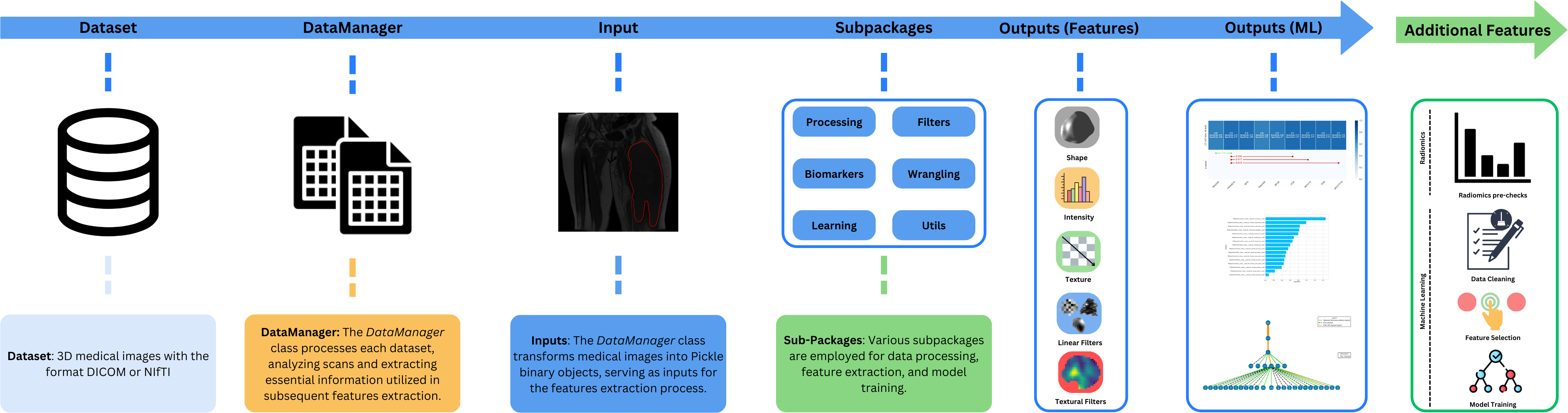
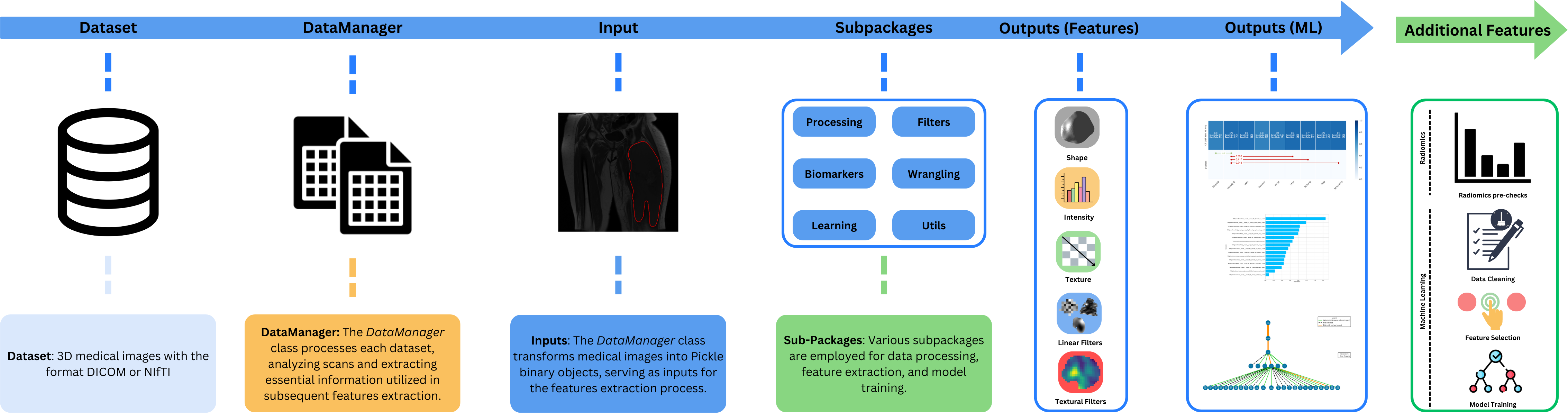
MEDimage is an open-source Python package that can be used for processing multi-modal medical images (MRI, CT or PET) and for extracting their radiomic features. This package is meant to facilitate the processing of medical images and the subsequent computation of all types of radiomic features while maintaining the reproducibility of analyses. This package has been standardized with the [IBSI](https://theibsi.github.io/) norms.
-
+
## 2. Installation
### Python installation
-The MEDimage package requires *Python 3.8* or more. If you don't have it installed on your machine, follow the instructions [here](https://github.com/MEDomics-UdeS/MEDimage/blob/main/python.md) to install it.
+The MEDimage package requires *Python 3.8* or more. If you don't have it installed on your machine, follow the instructions [here](https://github.com/MahdiAll99/MEDimage/blob/main/python.md) to install it.
### Package installation
You can easily install the ``MEDimage`` package from PyPI using:
@@ -99,22 +99,22 @@ med_obj.save_radiomics(
## 5. Tutorials
-We have created many [tutorial notebooks](https://github.com/MEDomics-UdeS/MEDimage/tree/main/notebooks) to assist you in learning how to use the different parts of the package. More details can be found in the [documentation](https://medimage.readthedocs.io/en/latest/tutorials.html).
+We have created many [tutorial notebooks](https://github.com/MahdiAll99/MEDimage/tree/main/notebooks) to assist you in learning how to use the different parts of the package. More details can be found in the [documentation](https://medimage.readthedocs.io/en/latest/tutorials.html).
## 6. IBSI Standardization
The image biomarker standardization initiative ([IBSI](https://theibsi.github.io)) is an independent international collaboration that aims to standardize the extraction of image biomarkers from acquired imaging. The IBSI therefore seeks to provide image biomarker nomenclature and definitions, benchmark datasets, and benchmark values to verify image processing and image biomarker calculations, as well as reporting guidelines, for high-throughput image analysis. We participate in this collaboration with our package to make sure it respects international nomenclatures and definitions. The participation was separated into two chapters:
- ### IBSI Chapter 1
- [The IBSI chapter 1](https://theibsi.github.io/ibsi1/) is dedicated to the standardization of commonly used radiomic features. It was initiated in September 2016 and reached completion in March 2020. We have created two [jupyter notebooks](https://github.com/MEDomics-UdeS/MEDimage/tree/main/notebooks/ibsi) for each phase of the chapter and made them available for the users to run the IBSI tests for themselves. The tests can also be explored in interactive Colab notebooks that are directly accessible here:
+ [The IBSI chapter 1](https://theibsi.github.io/ibsi1/) is dedicated to the standardization of commonly used radiomic features. It was initiated in September 2016 and reached completion in March 2020. We have created two [jupyter notebooks](https://github.com/MahdiAll99/MEDimage/tree/main/notebooks/ibsi) for each phase of the chapter and made them available for the users to run the IBSI tests for themselves. The tests can also be explored in interactive Colab notebooks that are directly accessible here:
- - **Phase 1**: [](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/ibsi/ibsi1p1.ipynb)
- - **Phase 2**: [](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/ibsi/ibsi1p2.ipynb)
+ - **Phase 1**: [](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/ibsi/ibsi1p1.ipynb)
+ - **Phase 2**: [](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/ibsi/ibsi1p2.ipynb)
- ### IBSI Chapter 2
- [The IBSI chapter 2](https://theibsi.github.io/ibsi2/) was launched in June 2020 and reached completion in February 2024. It is dedicated to the standardization of commonly used imaging filters in radiomic studies. We have created two [jupyter notebooks](https://github.com/MEDomics-UdeS/MEDimage/tree/main/notebooks/ibsi) for each phase of the chapter and made them available for the users to run the IBSI tests for themselves and validate image filtering and image biomarker calculations from filter response maps. The tests can also be explored in interactive Colab notebooks that are directly accessible here:
+ [The IBSI chapter 2](https://theibsi.github.io/ibsi2/) was launched in June 2020 and reached completion in February 2024. It is dedicated to the standardization of commonly used imaging filters in radiomic studies. We have created two [jupyter notebooks](https://github.com/MahdiAll99/MEDimage/tree/main/notebooks/ibsi) for each phase of the chapter and made them available for the users to run the IBSI tests for themselves and validate image filtering and image biomarker calculations from filter response maps. The tests can also be explored in interactive Colab notebooks that are directly accessible here:
- - **Phase 1**: [](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/ibsi/ibsi2p1.ipynb)
- - **Phase 2**: [](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/ibsi/ibsi2p2.ipynb)
+ - **Phase 1**: [](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/ibsi/ibsi2p1.ipynb)
+ - **Phase 2**: [](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/ibsi/ibsi2p2.ipynb)
Our team at *UdeS* (a.k.a. Université de Sherbrooke) has already submitted the benchmarked values to the [IBSI uploading website](https://ibsi.radiomics.hevs.ch/).
diff --git a/docs/extraction_config.rst b/docs/extraction_config.rst
index 0b4f22f..376e438 100644
--- a/docs/extraction_config.rst
+++ b/docs/extraction_config.rst
@@ -372,7 +372,7 @@ e.g.
{
"$schema": "http://json-schema.org/draft-04/schema#",
- "title": "discretisation",
+ "title": "discretization",
"description": "Discretization parameters.",
"type": "dict",
"options": {
@@ -443,7 +443,7 @@ e.g.
"type": "dict",
"options": {
"type": {
- "description": "List of discretisation algorithms: ``\"FBS\"`` for fixed bin size and
+ "description": "List of discretization algorithms: ``\"FBS\"`` for fixed bin size and
``\"FBN\"`` for fixed bin number. Texture features will be computed for each algorithm in the list",
"type": "List[string]"
},
@@ -461,7 +461,7 @@ e.g. for CT only (the parameters are the same for MR and PET):
{
"imParamCT" : {
- "discretisation" : {
+ "discretization" : {
"IH" : {
"type" : "FBS",
"val" : 25
diff --git a/docs/figures/MEDimageLogo.png b/docs/figures/MEDimageLogo.png
index 744652b..e610eee 100644
Binary files a/docs/figures/MEDimageLogo.png and b/docs/figures/MEDimageLogo.png differ
diff --git a/docs/processing.rst b/docs/processing.rst
index b256f05..b8ffe72 100644
--- a/docs/processing.rst
+++ b/docs/processing.rst
@@ -10,7 +10,7 @@ compute\_suv\_map
:undoc-members:
:show-inheritance:
-discretisation
+discretization
-----------------------------------------
.. automodule:: MEDimage.processing.discretisation
diff --git a/environment.yml b/environment.yml
index 430bdbf..02d97a5 100644
--- a/environment.yml
+++ b/environment.yml
@@ -8,7 +8,7 @@ dependencies:
- nibabel
- pandas<2.0.0
- pillow
- - pydicom>1.2.0, <=1.3.0
+ - pydicom
- pywavelets
- scikit-image
- scipy
diff --git a/pyproject.toml b/pyproject.toml
index 46ab7a7..0b62d96 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -1,6 +1,6 @@
[tool.poetry]
name = "medimage-pkg"
-version = "0.9.4"
+version = "0.9.7"
description = "MEDimage is a Python package for processing and extracting features from medical images"
authors = ["MEDomics Consortium "]
license = "GPL-3.0"
@@ -30,7 +30,7 @@ pandas = "<2.0.0"
Pillow = "*"
protobuf = "*"
pycaret = "*"
-pydicom = ">1.2.0, <=1.3.0"
+pydicom = "*"
PyWavelets = "*"
ray = { version = "*", extras = ["default"] }
scikit_image = "*"
diff --git a/requirements.txt b/requirements.txt
index 23b75a0..e3cbf27 100644
--- a/requirements.txt
+++ b/requirements.txt
@@ -13,7 +13,7 @@ pandas<2.0.0
Pillow
protobuf
pycaret
-pydicom>1.2.0, <=1.3.0
+pydicom
PyWavelets
ray[default]
scikit_image
diff --git a/setup.py b/setup.py
index 917e45e..4aa657d 100644
--- a/setup.py
+++ b/setup.py
@@ -14,7 +14,7 @@
setup(
name="MEDimage",
- version="0.9.4",
+ version="0.9.7",
author="MEDomics consortium",
author_email="medomics.info@gmail.com",
description="Python Open-source package for medical images processing and radiomic features extraction",
[](https://www.python.org/downloads/release/python-380/)
-[](https://test.pypi.org/project/medimage-pkg/0.2.0/)
-[](https://github.com/MEDomics-UdeS/MEDimage/actions/workflows/python-app.yml)
+[](https://pypi.org/project/medimage-pkg/)
+[](https://github.com/MahdiAll99/MEDimage/actions/workflows/python-app.yml)
[](https://medimage.readthedocs.io/en/latest/?badge=latest)
[](LICENSE)
-[](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/tutorial/DataManager-Tutorial.ipynb)
+[](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/tutorial/DataManager-Tutorial.ipynb)
@@ -27,13 +27,13 @@
## 1. Introduction
MEDimage is an open-source Python package that can be used for processing multi-modal medical images (MRI, CT or PET) and for extracting their radiomic features. This package is meant to facilitate the processing of medical images and the subsequent computation of all types of radiomic features while maintaining the reproducibility of analyses. This package has been standardized with the [IBSI](https://theibsi.github.io/) norms.
-
+
## 2. Installation
### Python installation
-The MEDimage package requires *Python 3.8* or more. If you don't have it installed on your machine, follow the instructions [here](https://github.com/MEDomics-UdeS/MEDimage/blob/main/python.md) to install it.
+The MEDimage package requires *Python 3.8* or more. If you don't have it installed on your machine, follow the instructions [here](https://github.com/MahdiAll99/MEDimage/blob/main/python.md) to install it.
### Package installation
You can easily install the ``MEDimage`` package from PyPI using:
@@ -99,22 +99,22 @@ med_obj.save_radiomics(
## 5. Tutorials
-We have created many [tutorial notebooks](https://github.com/MEDomics-UdeS/MEDimage/tree/main/notebooks) to assist you in learning how to use the different parts of the package. More details can be found in the [documentation](https://medimage.readthedocs.io/en/latest/tutorials.html).
+We have created many [tutorial notebooks](https://github.com/MahdiAll99/MEDimage/tree/main/notebooks) to assist you in learning how to use the different parts of the package. More details can be found in the [documentation](https://medimage.readthedocs.io/en/latest/tutorials.html).
## 6. IBSI Standardization
The image biomarker standardization initiative ([IBSI](https://theibsi.github.io)) is an independent international collaboration that aims to standardize the extraction of image biomarkers from acquired imaging. The IBSI therefore seeks to provide image biomarker nomenclature and definitions, benchmark datasets, and benchmark values to verify image processing and image biomarker calculations, as well as reporting guidelines, for high-throughput image analysis. We participate in this collaboration with our package to make sure it respects international nomenclatures and definitions. The participation was separated into two chapters:
- ### IBSI Chapter 1
- [The IBSI chapter 1](https://theibsi.github.io/ibsi1/) is dedicated to the standardization of commonly used radiomic features. It was initiated in September 2016 and reached completion in March 2020. We have created two [jupyter notebooks](https://github.com/MEDomics-UdeS/MEDimage/tree/main/notebooks/ibsi) for each phase of the chapter and made them available for the users to run the IBSI tests for themselves. The tests can also be explored in interactive Colab notebooks that are directly accessible here:
+ [The IBSI chapter 1](https://theibsi.github.io/ibsi1/) is dedicated to the standardization of commonly used radiomic features. It was initiated in September 2016 and reached completion in March 2020. We have created two [jupyter notebooks](https://github.com/MahdiAll99/MEDimage/tree/main/notebooks/ibsi) for each phase of the chapter and made them available for the users to run the IBSI tests for themselves. The tests can also be explored in interactive Colab notebooks that are directly accessible here:
- - **Phase 1**: [](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/ibsi/ibsi1p1.ipynb)
- - **Phase 2**: [](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/ibsi/ibsi1p2.ipynb)
+ - **Phase 1**: [](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/ibsi/ibsi1p1.ipynb)
+ - **Phase 2**: [](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/ibsi/ibsi1p2.ipynb)
- ### IBSI Chapter 2
- [The IBSI chapter 2](https://theibsi.github.io/ibsi2/) was launched in June 2020 and reached completion in February 2024. It is dedicated to the standardization of commonly used imaging filters in radiomic studies. We have created two [jupyter notebooks](https://github.com/MEDomics-UdeS/MEDimage/tree/main/notebooks/ibsi) for each phase of the chapter and made them available for the users to run the IBSI tests for themselves and validate image filtering and image biomarker calculations from filter response maps. The tests can also be explored in interactive Colab notebooks that are directly accessible here:
+ [The IBSI chapter 2](https://theibsi.github.io/ibsi2/) was launched in June 2020 and reached completion in February 2024. It is dedicated to the standardization of commonly used imaging filters in radiomic studies. We have created two [jupyter notebooks](https://github.com/MahdiAll99/MEDimage/tree/main/notebooks/ibsi) for each phase of the chapter and made them available for the users to run the IBSI tests for themselves and validate image filtering and image biomarker calculations from filter response maps. The tests can also be explored in interactive Colab notebooks that are directly accessible here:
- - **Phase 1**: [](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/ibsi/ibsi2p1.ipynb)
- - **Phase 2**: [](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/ibsi/ibsi2p2.ipynb)
+ - **Phase 1**: [](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/ibsi/ibsi2p1.ipynb)
+ - **Phase 2**: [](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/ibsi/ibsi2p2.ipynb)
Our team at *UdeS* (a.k.a. Université de Sherbrooke) has already submitted the benchmarked values to the [IBSI uploading website](https://ibsi.radiomics.hevs.ch/).
diff --git a/docs/extraction_config.rst b/docs/extraction_config.rst
index 0b4f22f..376e438 100644
--- a/docs/extraction_config.rst
+++ b/docs/extraction_config.rst
@@ -372,7 +372,7 @@ e.g.
{
"$schema": "http://json-schema.org/draft-04/schema#",
- "title": "discretisation",
+ "title": "discretization",
"description": "Discretization parameters.",
"type": "dict",
"options": {
@@ -443,7 +443,7 @@ e.g.
"type": "dict",
"options": {
"type": {
- "description": "List of discretisation algorithms: ``\"FBS\"`` for fixed bin size and
+ "description": "List of discretization algorithms: ``\"FBS\"`` for fixed bin size and
``\"FBN\"`` for fixed bin number. Texture features will be computed for each algorithm in the list",
"type": "List[string]"
},
@@ -461,7 +461,7 @@ e.g. for CT only (the parameters are the same for MR and PET):
{
"imParamCT" : {
- "discretisation" : {
+ "discretization" : {
"IH" : {
"type" : "FBS",
"val" : 25
diff --git a/docs/figures/MEDimageLogo.png b/docs/figures/MEDimageLogo.png
index 744652b..e610eee 100644
Binary files a/docs/figures/MEDimageLogo.png and b/docs/figures/MEDimageLogo.png differ
diff --git a/docs/processing.rst b/docs/processing.rst
index b256f05..b8ffe72 100644
--- a/docs/processing.rst
+++ b/docs/processing.rst
@@ -10,7 +10,7 @@ compute\_suv\_map
:undoc-members:
:show-inheritance:
-discretisation
+discretization
-----------------------------------------
.. automodule:: MEDimage.processing.discretisation
diff --git a/environment.yml b/environment.yml
index 430bdbf..02d97a5 100644
--- a/environment.yml
+++ b/environment.yml
@@ -8,7 +8,7 @@ dependencies:
- nibabel
- pandas<2.0.0
- pillow
- - pydicom>1.2.0, <=1.3.0
+ - pydicom
- pywavelets
- scikit-image
- scipy
diff --git a/pyproject.toml b/pyproject.toml
index 46ab7a7..0b62d96 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -1,6 +1,6 @@
[tool.poetry]
name = "medimage-pkg"
-version = "0.9.4"
+version = "0.9.7"
description = "MEDimage is a Python package for processing and extracting features from medical images"
authors = ["MEDomics Consortium "]
license = "GPL-3.0"
@@ -30,7 +30,7 @@ pandas = "<2.0.0"
Pillow = "*"
protobuf = "*"
pycaret = "*"
-pydicom = ">1.2.0, <=1.3.0"
+pydicom = "*"
PyWavelets = "*"
ray = { version = "*", extras = ["default"] }
scikit_image = "*"
diff --git a/requirements.txt b/requirements.txt
index 23b75a0..e3cbf27 100644
--- a/requirements.txt
+++ b/requirements.txt
@@ -13,7 +13,7 @@ pandas<2.0.0
Pillow
protobuf
pycaret
-pydicom>1.2.0, <=1.3.0
+pydicom
PyWavelets
ray[default]
scikit_image
diff --git a/setup.py b/setup.py
index 917e45e..4aa657d 100644
--- a/setup.py
+++ b/setup.py
@@ -14,7 +14,7 @@
setup(
name="MEDimage",
- version="0.9.4",
+ version="0.9.7",
author="MEDomics consortium",
author_email="medomics.info@gmail.com",
description="Python Open-source package for medical images processing and radiomic features extraction",
 [](https://www.python.org/downloads/release/python-380/)
-[](https://test.pypi.org/project/medimage-pkg/0.2.0/)
-[](https://github.com/MEDomics-UdeS/MEDimage/actions/workflows/python-app.yml)
+[](https://pypi.org/project/medimage-pkg/)
+[](https://github.com/MahdiAll99/MEDimage/actions/workflows/python-app.yml)
[](https://medimage.readthedocs.io/en/latest/?badge=latest)
[](LICENSE)
-[](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/tutorial/DataManager-Tutorial.ipynb)
+[](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/tutorial/DataManager-Tutorial.ipynb)
@@ -27,13 +27,13 @@
## 1. Introduction
MEDimage is an open-source Python package that can be used for processing multi-modal medical images (MRI, CT or PET) and for extracting their radiomic features. This package is meant to facilitate the processing of medical images and the subsequent computation of all types of radiomic features while maintaining the reproducibility of analyses. This package has been standardized with the [IBSI](https://theibsi.github.io/) norms.
-
+
## 2. Installation
### Python installation
-The MEDimage package requires *Python 3.8* or more. If you don't have it installed on your machine, follow the instructions [here](https://github.com/MEDomics-UdeS/MEDimage/blob/main/python.md) to install it.
+The MEDimage package requires *Python 3.8* or more. If you don't have it installed on your machine, follow the instructions [here](https://github.com/MahdiAll99/MEDimage/blob/main/python.md) to install it.
### Package installation
You can easily install the ``MEDimage`` package from PyPI using:
@@ -99,22 +99,22 @@ med_obj.save_radiomics(
## 5. Tutorials
-We have created many [tutorial notebooks](https://github.com/MEDomics-UdeS/MEDimage/tree/main/notebooks) to assist you in learning how to use the different parts of the package. More details can be found in the [documentation](https://medimage.readthedocs.io/en/latest/tutorials.html).
+We have created many [tutorial notebooks](https://github.com/MahdiAll99/MEDimage/tree/main/notebooks) to assist you in learning how to use the different parts of the package. More details can be found in the [documentation](https://medimage.readthedocs.io/en/latest/tutorials.html).
## 6. IBSI Standardization
The image biomarker standardization initiative ([IBSI](https://theibsi.github.io)) is an independent international collaboration that aims to standardize the extraction of image biomarkers from acquired imaging. The IBSI therefore seeks to provide image biomarker nomenclature and definitions, benchmark datasets, and benchmark values to verify image processing and image biomarker calculations, as well as reporting guidelines, for high-throughput image analysis. We participate in this collaboration with our package to make sure it respects international nomenclatures and definitions. The participation was separated into two chapters:
- ### IBSI Chapter 1
- [The IBSI chapter 1](https://theibsi.github.io/ibsi1/) is dedicated to the standardization of commonly used radiomic features. It was initiated in September 2016 and reached completion in March 2020. We have created two [jupyter notebooks](https://github.com/MEDomics-UdeS/MEDimage/tree/main/notebooks/ibsi) for each phase of the chapter and made them available for the users to run the IBSI tests for themselves. The tests can also be explored in interactive Colab notebooks that are directly accessible here:
+ [The IBSI chapter 1](https://theibsi.github.io/ibsi1/) is dedicated to the standardization of commonly used radiomic features. It was initiated in September 2016 and reached completion in March 2020. We have created two [jupyter notebooks](https://github.com/MahdiAll99/MEDimage/tree/main/notebooks/ibsi) for each phase of the chapter and made them available for the users to run the IBSI tests for themselves. The tests can also be explored in interactive Colab notebooks that are directly accessible here:
- - **Phase 1**: [](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/ibsi/ibsi1p1.ipynb)
- - **Phase 2**: [](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/ibsi/ibsi1p2.ipynb)
+ - **Phase 1**: [](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/ibsi/ibsi1p1.ipynb)
+ - **Phase 2**: [](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/ibsi/ibsi1p2.ipynb)
- ### IBSI Chapter 2
- [The IBSI chapter 2](https://theibsi.github.io/ibsi2/) was launched in June 2020 and reached completion in February 2024. It is dedicated to the standardization of commonly used imaging filters in radiomic studies. We have created two [jupyter notebooks](https://github.com/MEDomics-UdeS/MEDimage/tree/main/notebooks/ibsi) for each phase of the chapter and made them available for the users to run the IBSI tests for themselves and validate image filtering and image biomarker calculations from filter response maps. The tests can also be explored in interactive Colab notebooks that are directly accessible here:
+ [The IBSI chapter 2](https://theibsi.github.io/ibsi2/) was launched in June 2020 and reached completion in February 2024. It is dedicated to the standardization of commonly used imaging filters in radiomic studies. We have created two [jupyter notebooks](https://github.com/MahdiAll99/MEDimage/tree/main/notebooks/ibsi) for each phase of the chapter and made them available for the users to run the IBSI tests for themselves and validate image filtering and image biomarker calculations from filter response maps. The tests can also be explored in interactive Colab notebooks that are directly accessible here:
- - **Phase 1**: [](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/ibsi/ibsi2p1.ipynb)
- - **Phase 2**: [](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/ibsi/ibsi2p2.ipynb)
+ - **Phase 1**: [](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/ibsi/ibsi2p1.ipynb)
+ - **Phase 2**: [](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/ibsi/ibsi2p2.ipynb)
Our team at *UdeS* (a.k.a. Université de Sherbrooke) has already submitted the benchmarked values to the [IBSI uploading website](https://ibsi.radiomics.hevs.ch/).
diff --git a/docs/extraction_config.rst b/docs/extraction_config.rst
index 0b4f22f..376e438 100644
--- a/docs/extraction_config.rst
+++ b/docs/extraction_config.rst
@@ -372,7 +372,7 @@ e.g.
{
"$schema": "http://json-schema.org/draft-04/schema#",
- "title": "discretisation",
+ "title": "discretization",
"description": "Discretization parameters.",
"type": "dict",
"options": {
@@ -443,7 +443,7 @@ e.g.
"type": "dict",
"options": {
"type": {
- "description": "List of discretisation algorithms: ``\"FBS\"`` for fixed bin size and
+ "description": "List of discretization algorithms: ``\"FBS\"`` for fixed bin size and
``\"FBN\"`` for fixed bin number. Texture features will be computed for each algorithm in the list",
"type": "List[string]"
},
@@ -461,7 +461,7 @@ e.g. for CT only (the parameters are the same for MR and PET):
{
"imParamCT" : {
- "discretisation" : {
+ "discretization" : {
"IH" : {
"type" : "FBS",
"val" : 25
diff --git a/docs/figures/MEDimageLogo.png b/docs/figures/MEDimageLogo.png
index 744652b..e610eee 100644
Binary files a/docs/figures/MEDimageLogo.png and b/docs/figures/MEDimageLogo.png differ
diff --git a/docs/processing.rst b/docs/processing.rst
index b256f05..b8ffe72 100644
--- a/docs/processing.rst
+++ b/docs/processing.rst
@@ -10,7 +10,7 @@ compute\_suv\_map
:undoc-members:
:show-inheritance:
-discretisation
+discretization
-----------------------------------------
.. automodule:: MEDimage.processing.discretisation
diff --git a/environment.yml b/environment.yml
index 430bdbf..02d97a5 100644
--- a/environment.yml
+++ b/environment.yml
@@ -8,7 +8,7 @@ dependencies:
- nibabel
- pandas<2.0.0
- pillow
- - pydicom>1.2.0, <=1.3.0
+ - pydicom
- pywavelets
- scikit-image
- scipy
diff --git a/pyproject.toml b/pyproject.toml
index 46ab7a7..0b62d96 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -1,6 +1,6 @@
[tool.poetry]
name = "medimage-pkg"
-version = "0.9.4"
+version = "0.9.7"
description = "MEDimage is a Python package for processing and extracting features from medical images"
authors = ["MEDomics Consortium
[](https://www.python.org/downloads/release/python-380/)
-[](https://test.pypi.org/project/medimage-pkg/0.2.0/)
-[](https://github.com/MEDomics-UdeS/MEDimage/actions/workflows/python-app.yml)
+[](https://pypi.org/project/medimage-pkg/)
+[](https://github.com/MahdiAll99/MEDimage/actions/workflows/python-app.yml)
[](https://medimage.readthedocs.io/en/latest/?badge=latest)
[](LICENSE)
-[](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/tutorial/DataManager-Tutorial.ipynb)
+[](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/tutorial/DataManager-Tutorial.ipynb)
@@ -27,13 +27,13 @@
## 1. Introduction
MEDimage is an open-source Python package that can be used for processing multi-modal medical images (MRI, CT or PET) and for extracting their radiomic features. This package is meant to facilitate the processing of medical images and the subsequent computation of all types of radiomic features while maintaining the reproducibility of analyses. This package has been standardized with the [IBSI](https://theibsi.github.io/) norms.
-
+
## 2. Installation
### Python installation
-The MEDimage package requires *Python 3.8* or more. If you don't have it installed on your machine, follow the instructions [here](https://github.com/MEDomics-UdeS/MEDimage/blob/main/python.md) to install it.
+The MEDimage package requires *Python 3.8* or more. If you don't have it installed on your machine, follow the instructions [here](https://github.com/MahdiAll99/MEDimage/blob/main/python.md) to install it.
### Package installation
You can easily install the ``MEDimage`` package from PyPI using:
@@ -99,22 +99,22 @@ med_obj.save_radiomics(
## 5. Tutorials
-We have created many [tutorial notebooks](https://github.com/MEDomics-UdeS/MEDimage/tree/main/notebooks) to assist you in learning how to use the different parts of the package. More details can be found in the [documentation](https://medimage.readthedocs.io/en/latest/tutorials.html).
+We have created many [tutorial notebooks](https://github.com/MahdiAll99/MEDimage/tree/main/notebooks) to assist you in learning how to use the different parts of the package. More details can be found in the [documentation](https://medimage.readthedocs.io/en/latest/tutorials.html).
## 6. IBSI Standardization
The image biomarker standardization initiative ([IBSI](https://theibsi.github.io)) is an independent international collaboration that aims to standardize the extraction of image biomarkers from acquired imaging. The IBSI therefore seeks to provide image biomarker nomenclature and definitions, benchmark datasets, and benchmark values to verify image processing and image biomarker calculations, as well as reporting guidelines, for high-throughput image analysis. We participate in this collaboration with our package to make sure it respects international nomenclatures and definitions. The participation was separated into two chapters:
- ### IBSI Chapter 1
- [The IBSI chapter 1](https://theibsi.github.io/ibsi1/) is dedicated to the standardization of commonly used radiomic features. It was initiated in September 2016 and reached completion in March 2020. We have created two [jupyter notebooks](https://github.com/MEDomics-UdeS/MEDimage/tree/main/notebooks/ibsi) for each phase of the chapter and made them available for the users to run the IBSI tests for themselves. The tests can also be explored in interactive Colab notebooks that are directly accessible here:
+ [The IBSI chapter 1](https://theibsi.github.io/ibsi1/) is dedicated to the standardization of commonly used radiomic features. It was initiated in September 2016 and reached completion in March 2020. We have created two [jupyter notebooks](https://github.com/MahdiAll99/MEDimage/tree/main/notebooks/ibsi) for each phase of the chapter and made them available for the users to run the IBSI tests for themselves. The tests can also be explored in interactive Colab notebooks that are directly accessible here:
- - **Phase 1**: [](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/ibsi/ibsi1p1.ipynb)
- - **Phase 2**: [](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/ibsi/ibsi1p2.ipynb)
+ - **Phase 1**: [](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/ibsi/ibsi1p1.ipynb)
+ - **Phase 2**: [](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/ibsi/ibsi1p2.ipynb)
- ### IBSI Chapter 2
- [The IBSI chapter 2](https://theibsi.github.io/ibsi2/) was launched in June 2020 and reached completion in February 2024. It is dedicated to the standardization of commonly used imaging filters in radiomic studies. We have created two [jupyter notebooks](https://github.com/MEDomics-UdeS/MEDimage/tree/main/notebooks/ibsi) for each phase of the chapter and made them available for the users to run the IBSI tests for themselves and validate image filtering and image biomarker calculations from filter response maps. The tests can also be explored in interactive Colab notebooks that are directly accessible here:
+ [The IBSI chapter 2](https://theibsi.github.io/ibsi2/) was launched in June 2020 and reached completion in February 2024. It is dedicated to the standardization of commonly used imaging filters in radiomic studies. We have created two [jupyter notebooks](https://github.com/MahdiAll99/MEDimage/tree/main/notebooks/ibsi) for each phase of the chapter and made them available for the users to run the IBSI tests for themselves and validate image filtering and image biomarker calculations from filter response maps. The tests can also be explored in interactive Colab notebooks that are directly accessible here:
- - **Phase 1**: [](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/ibsi/ibsi2p1.ipynb)
- - **Phase 2**: [](https://colab.research.google.com/github/MEDomics-UdeS/MEDimage/blob/main/notebooks/ibsi/ibsi2p2.ipynb)
+ - **Phase 1**: [](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/ibsi/ibsi2p1.ipynb)
+ - **Phase 2**: [](https://colab.research.google.com/github/MahdiAll99/MEDimage/blob/main/notebooks/ibsi/ibsi2p2.ipynb)
Our team at *UdeS* (a.k.a. Université de Sherbrooke) has already submitted the benchmarked values to the [IBSI uploading website](https://ibsi.radiomics.hevs.ch/).
diff --git a/docs/extraction_config.rst b/docs/extraction_config.rst
index 0b4f22f..376e438 100644
--- a/docs/extraction_config.rst
+++ b/docs/extraction_config.rst
@@ -372,7 +372,7 @@ e.g.
{
"$schema": "http://json-schema.org/draft-04/schema#",
- "title": "discretisation",
+ "title": "discretization",
"description": "Discretization parameters.",
"type": "dict",
"options": {
@@ -443,7 +443,7 @@ e.g.
"type": "dict",
"options": {
"type": {
- "description": "List of discretisation algorithms: ``\"FBS\"`` for fixed bin size and
+ "description": "List of discretization algorithms: ``\"FBS\"`` for fixed bin size and
``\"FBN\"`` for fixed bin number. Texture features will be computed for each algorithm in the list",
"type": "List[string]"
},
@@ -461,7 +461,7 @@ e.g. for CT only (the parameters are the same for MR and PET):
{
"imParamCT" : {
- "discretisation" : {
+ "discretization" : {
"IH" : {
"type" : "FBS",
"val" : 25
diff --git a/docs/figures/MEDimageLogo.png b/docs/figures/MEDimageLogo.png
index 744652b..e610eee 100644
Binary files a/docs/figures/MEDimageLogo.png and b/docs/figures/MEDimageLogo.png differ
diff --git a/docs/processing.rst b/docs/processing.rst
index b256f05..b8ffe72 100644
--- a/docs/processing.rst
+++ b/docs/processing.rst
@@ -10,7 +10,7 @@ compute\_suv\_map
:undoc-members:
:show-inheritance:
-discretisation
+discretization
-----------------------------------------
.. automodule:: MEDimage.processing.discretisation
diff --git a/environment.yml b/environment.yml
index 430bdbf..02d97a5 100644
--- a/environment.yml
+++ b/environment.yml
@@ -8,7 +8,7 @@ dependencies:
- nibabel
- pandas<2.0.0
- pillow
- - pydicom>1.2.0, <=1.3.0
+ - pydicom
- pywavelets
- scikit-image
- scipy
diff --git a/pyproject.toml b/pyproject.toml
index 46ab7a7..0b62d96 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -1,6 +1,6 @@
[tool.poetry]
name = "medimage-pkg"
-version = "0.9.4"
+version = "0.9.7"
description = "MEDimage is a Python package for processing and extracting features from medical images"
authors = ["MEDomics Consortium