-
Notifications
You must be signed in to change notification settings - Fork 1
/
07-reporting.Rmd
51 lines (30 loc) · 3.03 KB
/
07-reporting.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
# Reporting results
Once we have done an enrichment analysis - how to communicate the results? Like everything - it depends what you need to say. Some examples are below.
## In text
If you just need to emphasise that the differential expression relates to a condition of interest, you don't need much:
> Genes differentially expressed after SH-SY5Y cell differentiation were enriched for GO term "regulation of neurogenesis", adjusted pval < 0.01).
And in the methods:
> Enrichment calculated for differentially expressed genes with the g:GOSt enrichment tool [(Raudvere et al, 2019)](https://academic.oup.com/nar/article/47/W1/W191/5486750) using a background of tested genes.
## As a table
For a more complete view, a table of the significant or top _n_ terms can be useful. E.g In a supplementary figure
The top 10 enriched GO terms for the differentially expressed genes;
| GO:BP Term | Term ID | Adjusted p-value | Term Size | Num DE Genes |
|------------------------------------------|------------|-----------------:|----------:|------------------:|
| system process | GO:0003008 | 2.90E-04 | 1243 | 45 |
| nervous system development | GO:0007399 | 4.19E-04 | 1985 | 60 |
| regulation of cell development | GO:0060284 | 1.98E-03 | 795 | 33 |
| central nervous system development | GO:0007417 | 2.67E-03 | 805 | 33 |
| regulation of neurogenesis | GO:0050767 | 5.59E-03 | 707 | 30 |
| regulation of nervous system development | GO:0051960 | 5.78E-03 | 790 | 32 |
| regulation of cell differentiation | GO:0045595 | 6.03E-03 | 1471 | 47 |
| multicellular organismal process | GO:0032501 | 2.04E-02 | 5300 | 111 |
| system development | GO:0048731 | 2.88E-02 | 3668 | 85 |
| neurogenesis | GO:0022008 | 4.36E-02 | 1375 | 43 |
## As a figure
Plots of -log p-value are a popular option for a figure. By taking the exponent of the p-value bigger bars indicate more significance.
For instance, this figure is taken from the [(Pezzini et al. 2017)](https://link.springer.com/article/10.1007%2Fs10571-016-0403-y) paper.
Note that in this figure shows 2 specifics categories from the IPA (ingenuity) database using the IPA tool (not covered here), so the terms themselves differ.
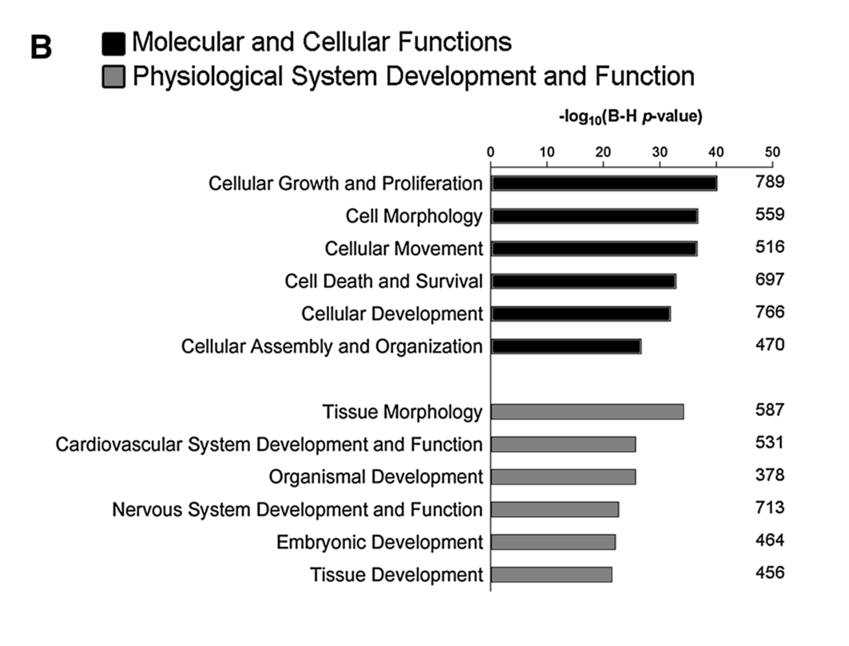
You can also build on such a plot with number of genes and the like. For an example, see the 'bubble' chart produced by a package called pathfindR here: https://www.biostars.org/p/322415/
----
There are also tools like cluego (a cytoscape plugin) that build enriched terms into a network : http://apps.cytoscape.org/apps/cluego