You signed in with another tab or window. Reload to refresh your session.You signed out in another tab or window. Reload to refresh your session.You switched accounts on another tab or window. Reload to refresh your session.Dismiss alert
Hi
Thanks to develop so wonderful tool, but I had some problems when I apply SNIPER models to my hic data.
When I have downloaded pre-computed SNIPER models, I found three kinds of files, autoencoder/encoder/classifier. I have no idea to select autoencoder or encoder, so I tried it all.
When I used autoencoder file, I got error as follows
ValueError: Error when checking input: expected dense_17_input to have shape (128,) but got array with shape (13393,)
When I used `encoder` file, I got warnings as follows
/dellstorage02/quj_lab/jiqianzhao/04_software/anaconda3/envs/SNIPER/lib/python3.6/site-packages/keras/engine/saving.py:292: UserWarning: No training configuration found in save file: the model was not compiled. Compile it manually
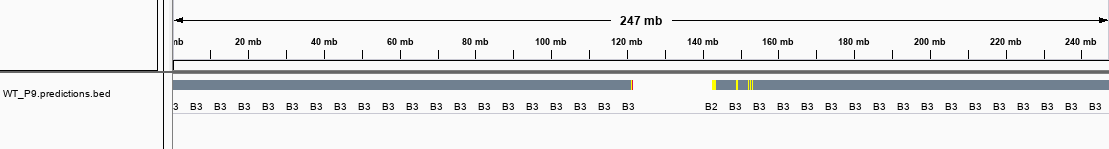
and its output bed files looks very strange. Its all "B3" subcompartments through genome.
Hi, your script inputs are correct. I suspect the ValueError is because the autoencoder outputs a high-dimensional vector and the classifier expects a low-dimensional input.
The all-B3 outputs you're seeing are likely because either:
The pre-computed model you selected expects much higher coverage than the hic file you've inputted.
Your inter-chromosomal contact map is almost completely devoid of signal. There's a good chance SNIPER will predict B3 across the board because B3 signals are generally associated with a lack of inter-chromosomal signal. Of the cell types we've tested, the inter-chromosomal signals of non-B3 subcompartments were sparse but still distinguishable.
Hi,
Thanks for your kind reply, its very helpful for me.
There are about 100 million interchromosomal contacts in my hic data, so I choose the pre-computed models from downsampled data (10%) , now I got the normal output.
But I have a few questions about the percentage of downsampled data. If I select one pre-computed model from lower coverage than my hic data, would SNIPER predict more incorrect non-B3 subcompartments, more false positives?
Looking forward your reply and thanks in advance!
Hi
Thanks to develop so wonderful tool, but I had some problems when I apply SNIPER models to my hic data.
When I have downloaded pre-computed SNIPER models, I found three kinds of files,
autoencoder/encoder/classifier. I have no idea to selectautoencoderorencoder, so I tried it all.When I used
autoencoderfile, I got error as followsand its output bed files looks very strange. Its all "B3" subcompartments through genome.
I have no idea to deal with it, Could you give me some sugesstions?
Thanks in advance!
Qianzhao
The text was updated successfully, but these errors were encountered: