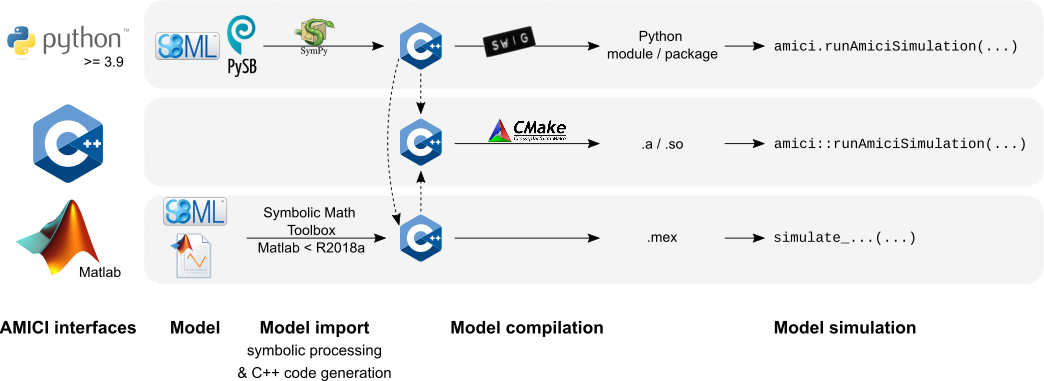
AMICI provides a multi-language (Python, C++, Matlab) interface for the
SUNDIALS solvers
CVODES
(for ordinary differential equations) and
IDAS
(for algebraic differential equations). AMICI allows the user to read
differential equation models specified as SBML
or PySB
and automatically compiles such models into Python modules, C++ libraries or
Matlab .mex simulation files.
In contrast to the (no longer maintained) sundialsTB Matlab interface, all necessary functions are transformed into native C++ code, which allows for a significantly faster simulation.
Beyond forward integration, the compiled simulation file also allows for forward sensitivity analysis, steady state sensitivity analysis and adjoint sensitivity analysis for likelihood-based output functions.
The interface was designed to provide routines for efficient gradient computation in parameter estimation of biochemical reaction models, but it is also applicable to a wider range of differential equation constrained optimization problems.
- SBML import
- PySB import
- Generation of C++ code for model simulation and sensitivity computation
- Access to and high customizability of CVODES and IDAS solver
- Python, C++, Matlab interface
- Sensitivity analysis
- forward
- steady state
- adjoint
- first- and second-order
- Pre-equilibration and pre-simulation conditions
- Support for discrete events and logical operations
The AMICI workflow starts with importing a model from either
SBML (Matlab, Python), PySB (Python),
or a Matlab definition of the model (Matlab-only). From this input,
all equations for model simulation
are derived symbolically and C++ code is generated. This code is then
compiled into a C++ library, a Python module, or a Matlab .mex file and
is then used for model simulation.
The AMICI source code is available at https://github.com/AMICI-dev/AMICI/. To install AMICI, first read the installation instructions for Python, C++ or Matlab. There are also instructions for using AMICI inside containers.
To get you started with Python-AMICI, the best way might be checking out this
Jupyter notebook
.
To get started with Matlab-AMICI, various examples are available in matlab/examples/.
Comprehensive documentation is available at https://amici.readthedocs.io/en/latest/.
Any contributions to AMICI are welcome (code, bug reports, suggestions for improvements, ...).
In case of questions or problems with using AMICI, feel free to post an issue on GitHub. We are trying to get back to you quickly.
There are several tools for parameter estimation offering good integration with AMICI:
- pyPESTO: Python library for optimization, sampling and uncertainty analysis
- pyABC: Python library for parallel and scalable ABC-SMC (Approximate Bayesian Computation - Sequential Monte Carlo)
- parPE: C++ library for parameter estimation of ODE models offering distributed memory parallelism with focus on problems with many simulation conditions.
Citeable DOI for the latest AMICI release:
There is a list of publications using AMICI. If you used AMICI in your work, we are happy to include your project, please let us know via a GitHub issue.
When using AMICI in your project, please cite
- Fröhlich, F., Weindl, D., Schälte, Y., Pathirana, D., Paszkowski, Ł., Lines, G.T., Stapor, P. and Hasenauer, J., 2021. AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models. Bioinformatics, btab227, DOI:10.1093/bioinformatics/btab227.
@article{frohlich2020amici,
title={AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models},
author={Fr{\"o}hlich, Fabian and Weindl, Daniel and Sch{\"a}lte, Yannik and Pathirana, Dilan and Paszkowski, {\L}ukasz and Lines, Glenn Terje and Stapor, Paul and Hasenauer, Jan},
journal = {Bioinformatics},
year = {2021},
month = {04},
issn = {1367-4803},
doi = {10.1093/bioinformatics/btab227},
note = {btab227},
eprint = {https://academic.oup.com/bioinformatics/advance-article-pdf/doi/10.1093/bioinformatics/btab227/36866220/btab227.pdf},
}
When presenting work that employs AMICI, feel free to use one of the icons in documentation/gfx/, which are available under a CC0 license: