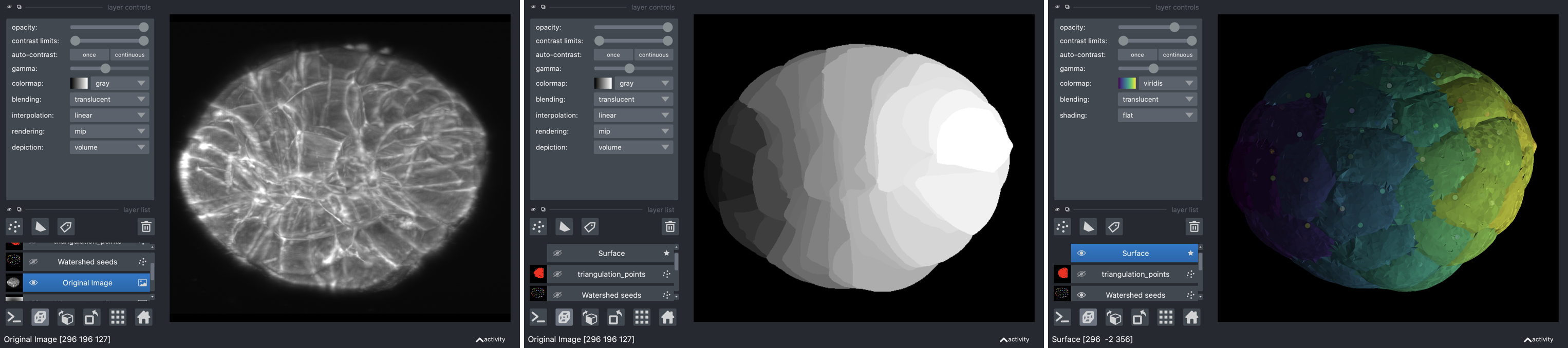
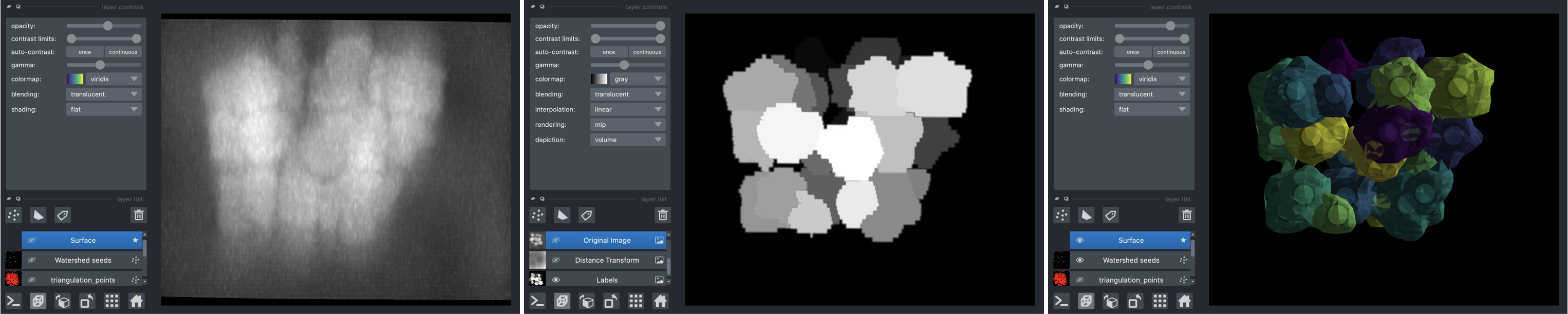
Delaunay-Watershed-3D is an algorithm designed to reconstruct in 3D a sparse surface mesh representation of the geometry of multicellular structures or nuclei from instance segmentations. It accomplishes this by building multimaterial meshes from segmentation masks. These multimaterial meshes are perfectly suited for storage, geometrical analysis, sharing and visualization of data. We provide as well visualization tools based on polyscope and napari.
Delaunay-Watershed was created by Sacha Ichbiah during his PhD in Turlier Lab, and is improved and maintained by Matthieu Perez and Hervé Turlier. For support, please open an issue. If you use this library in your work please cite the paper.
If you are interested in 2D images and meshes, please look at the foambryo-2D package instead.
Introductory notebooks are provided for two examples (cells or nuclei in multicellular aggregates). The algorithm takes as input 3D segmentation masks and returns multimaterial triangle meshes in 3D.
This method is used as a backend for foambryo, our 3D tension and pressure inference Python library.
We recommend to install delaunay-watershed from the PyPI repository directly, in a virtual environment.
pip install delaunay-watershed-3dIf you want to use our visualization tools, install the package with the viewing option:
pip install "delaunay-watershed-3d[viewing]"For developers, you may also install delaunay-watershed by cloning the source code and installing from the local directory
git clone https://github.com/VirtualEmbryo/delaunay-watershed.git
pip install pathtopackage/delaunay-watershedLoad an instance segmentation, construct its multimaterial mesh, save it to a file for later, and visualize it:
from dw3d import get_default_mesh_reconstruction_algorithm
from dw3D.viewing import plot_cells_polyscope
# Get a mesh reconstruction algorithm
mesh_reconstruction_algorithm = get_default_mesh_reconstruction_algorithm()
# Load the segmentation image
import skimage.io as io
segmentation_mask = io.imread("data/Images/1.tif")
# Reconstruct a multimaterial mesh from the mask using the mesh reconstruction algorithm
mesh_reconstruction_algorithm.construct_mesh_from_segmentation_mask(segmentation_mask)
# Save the last constructed mesh
mesh_reconstruction_algorithm.save_to_vtk_mesh("mesh_from_segmentation.vtk", binary_mode=True)
# Plot the last constructed mesh
plot_cells_polyscope(mesh_reconstruction_algorithm)Geometry can be analyzed later, in foambryo for example.
For more examples and documentation, see the notebooks:
There is also an advanced notebook if you want to tinkle with the algoritm: Advanced use.
See the notebook on mesh reconstruction and visualization.
Segmentation masks from Guignard et al.
See the notebook on mask compression and reconstruction.
Segmentation masks from Stardist
If you use this tool, please cite the associated paper. Do not hesitate to contact Matthieu Perez and Hervé Turlier for practical questions and applications. We hope that Delaunay-Watershed could help biologists and physicists to shed light on the mechanical aspects of early development.
@article {Ichbiah2023.04.12.536641,
author = {Sacha Ichbiah and Fabrice Delbary and Alex McDougall and R{\'e}mi Dumollard and Herv{\'e} Turlier},
title = {Embryo mechanics cartography: inference of 3D force atlases from fluorescence microscopy},
elocation-id = {2023.04.12.536641},
year = {2023},
doi = {10.1101/2023.04.12.536641},
publisher = {Cold Spring Harbor Laboratory},
abstract = {The morphogenesis of tissues and embryos results from a tight interplay between gene expression, biochemical signaling and mechanics. Although sequencing methods allow the generation of cell-resolved spatio-temporal maps of gene expression in developing tissues, creating similar maps of cell mechanics in 3D has remained a real challenge. Exploiting the foam-like geometry of cells in embryos, we propose a robust end-to-end computational method to infer spatiotemporal atlases of cellular forces from fluorescence microscopy images of cell membranes. Our method generates precise 3D meshes of cell geometry and successively predicts relative cell surface tensions and pressures in the tissue. We validate it with 3D active foam simulations, study its noise sensitivity, and prove its biological relevance in mouse, ascidian and C. elegans embryos. 3D inference allows us to recover mechanical features identified previously, but also predicts new ones, unveiling potential new insights on the spatiotemporal regulation of cell mechanics in early embryos. Our code is freely available and paves the way for unraveling the unknown mechanochemical feedbacks that control embryo and tissue morphogenesis.Competing Interest StatementThe authors have declared no competing interest.},
URL = {https://www.biorxiv.org/content/early/2023/04/13/2023.04.12.536641},
eprint = {https://www.biorxiv.org/content/early/2023/04/13/2023.04.12.536641.full.pdf},
journal = {bioRxiv}
}
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.