ObiWan-Microbi is a toolkit collection for semi-automated segmentation and annotation of (biological) image data in the cloud. It utilizes an OMERO image server for data magement. We contribute SegUI - a browser-based tool for semi-automated manual segmentation and annotation - and SegServe - a remote neural network execution server. All three components are combined into a docker-compose setup to be easily deployed on a local computer or in a cloud system.
- install docker
- install docker-compose
- recommended: CUDA capable GPU for accelerated deep-learning execution.
- recommended: linux system for hosting (tested on ubuntu)
ObiWan-Microbi can be installed on your local system, network servers or cloud setups. For small instances and testing, desktop computers or server computers are recommended. For large-scale setups, we recommend a cloud solution.
The installation is performed once on your system. Afterwards, you can access the services in your local network with any computer, notebook, tablet or smartphone.
Please download this repository including its submodules using
git clone --recurse-submodules https://github.com/hip-satomi/ObiWan-Microbi.git
cd ObiWan-MicrobiThe toolkit has to be built and launched using your bash shell
sudo docker-compose up --buildNote: Building will take a while for the first time (~1h) because all the necessary resources are downloaded, compiled and provided. However, this only has to be performed once: all following launches will be fast.
After the toolkit startup the defaults user (username: root, password: omero) is created and the following services are available:
- SegUI interface for annotation at http://localhost.
- SegServe interface for executing segmentation algorithms with the respective openapi documentation of the REST API http://localhost/segService/.
- OmeroWeb interface at http://localhost:4080.
- OMERO server at the default ports (4064) accessible with the default configuration of OMERO Insight.
Note: Please make sure that the ports 80, 4080 and 4064 are not used on your computer. Otherwise the launch of ObiWan-Microbi will fail.
Note: It is possible to connect ObiWan-Microbi to an existing OMERO server. If you would like to do so, please contact us.
We also provided a GPU tailored version of the toolkit to utilize the faster segmentation performance of Nvidia GPU hardware. Make sure that you have a very recent docker-compose version, an Nvidia driver installed and please install nvidia-docker2 to use your GPU in the docker container.
Now you can repeat the startup procedure with the GPU version
sudo docker-compose -f docker-compose.gpu.yml up --buildNote: GPU usage can dramatically speed up segmentation execution, especially for larger images (~20). Therfore, GPU usage is recommended in production usage. However, incompatibility problems between the computer NVIDIA driver and the NVIDIA driver used in the docker container can lead to problems. If you run into problems with your GPU please create an issue.
Installing ObiWan-Microbi on a Windows computer requires admin permissions and is possible using the Windows subsystem for Linux 2 (WSL2). Please follow the instructions here in order to get docker and Linux running on your Windows machine.
After that launch the linux distribution in WSL2 mode and proceed with the normal installation instructions.
We used Ubuntu 22.04.2 LTS in the WSL mode during our tests.
Why is Linux recommended?
ObiWan-Microbion Linux is much better supported, especially when GPU acceleration is needed for faster segmentation prediction. Therefore, the Linux installation is recommended. For initial testing, the Windows setup can be used.
For testing purposes, 5 simulated microbial time-lapse sequences are available after building the toolkit.
You can open the SegUI browsing view at http://localhost and login with default admin credentials (username: root, password: omero).
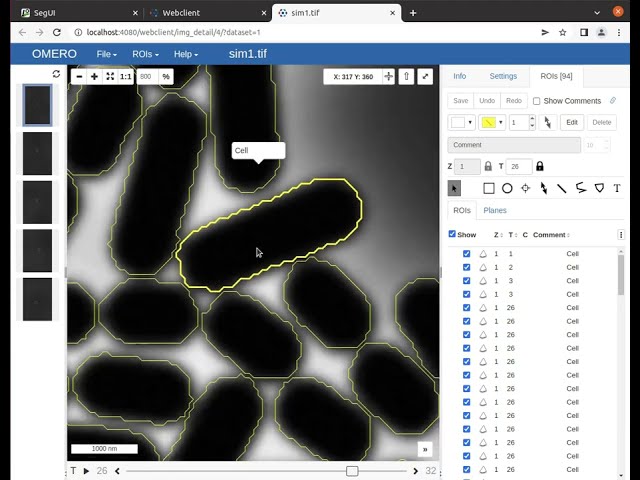
In this video tutorial, we show how to browse the data on your OMERO instance.
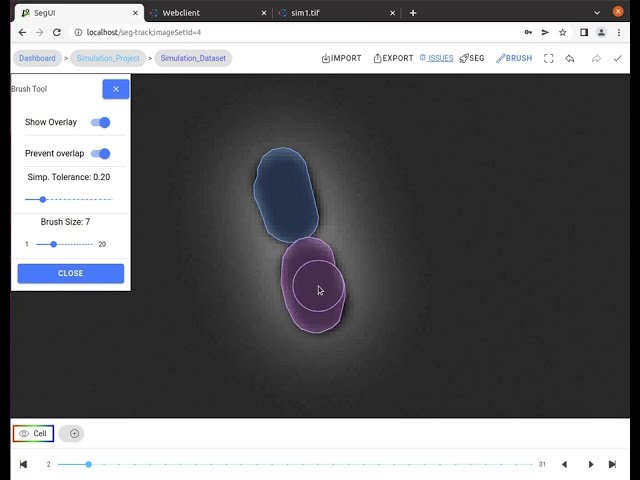
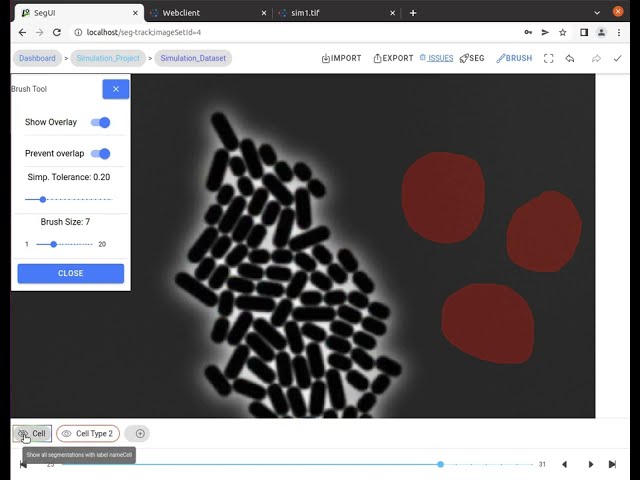
In this video tutorial, we show the segmentation features for fast manual cell segmentation.
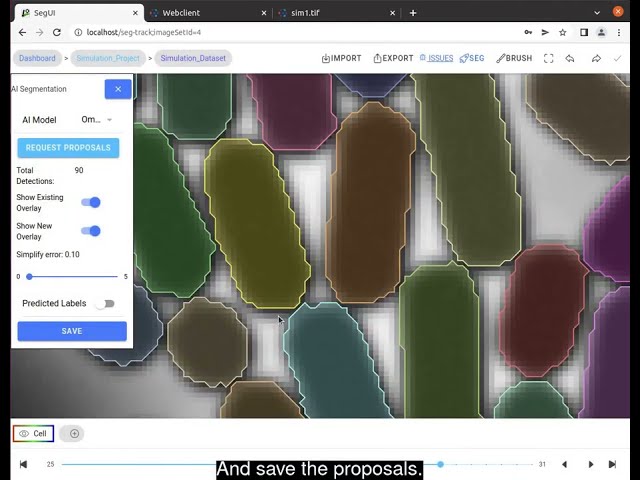
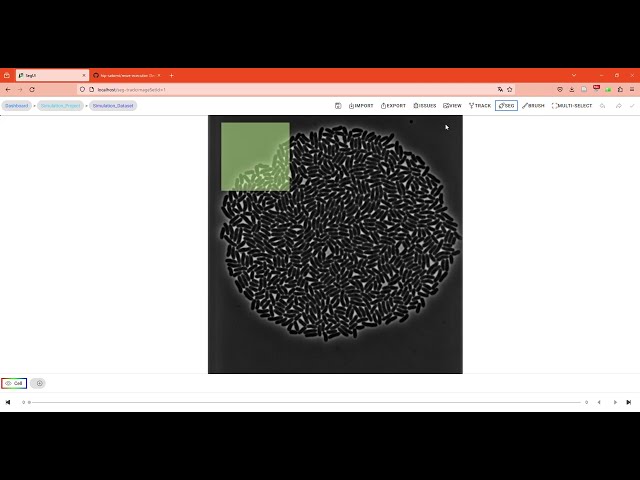
In this video tutorial, we demonstrate the remote execution of AI segmentation using the user interface.
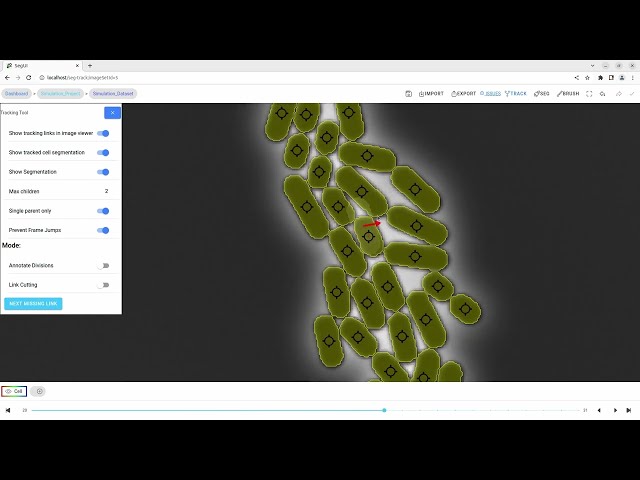
In this video tutorial, we show the manual tracking functionality.
In this vidoe tutorial, we show the import and export functionality into OMERO's native formats.
In this video tutorial, we demonstrate complex image annotation with multiple class labels.
In this video tutorial, we show how to add custom segmentation models and utilize them in the web interface.
If you want to train your own deep-learning segmentation to get high-quality results on your custom data, we highly recommend using microbeSEG. It allows to manage large training datasets, supports cropping to minimize annotation work, connects to ObiWan-Microbi toolkit for annotation and data management and allows to train microbial segmentation models. Check it out 🚀
| Key | Action |
|---|---|
| Left arrow | Previous image in the timelapse |
| Right arrow | Next Image in the timelapse |
| Key | Action |
|---|---|
| A/Enter | Add new segmentation annotation |
| Del | Delete selected segmentation |
| O | Show/Hide overlays |
| Key | Action |
|---|---|
| D | Hold the d key to select multiple children for a cell |
| C | Activate/Deactivate edge cutting mode |
In order to run ObiWan-Microbi with an existing OMERO instance you can edit the OMERO web endpoint here pointing to the correct web address. In that case, the omero containers can be remove from the docker-compose script. Furthermore, it is recommended to run ObiWan-Microbi on the same computer as OMERO to prevent problems with cross-site references.
A complete feature list of the ObiWan-Microbi software platform
- SegServe - Remote AI execution: Execute AI approaches on a single server with specialized hardware (e.g. GPU).
- Expandable AI Zoo: Segmentation approaches are packaged as git repositories and mflow and anaconda are used for dependency management. Installations are performed on demand. Your custom model only needs a git repo.
- Reproducibility: AI approaches are executed with version specification (e.g. current git commit hash) and can be reproduced at any time.
- State-of-the art microbial segmentation: Our model zoo currently contains the segmentation approaches Cellpose, Omnipose, Hybrid-Task-Cascade and general object detector Yolov5.
- Efficient resource usage: Multiple users access the same computation hardware increasing efficiency and utilization. At the same time specialized hardware (e.g. GPUs) are not necessary on end user devices.
- OpenAPI REST schema: SegServe can also be used by third-party applications via its REST interface. Its usage is automatically documented using the OpenAPI standard.
- User-friendly ground truth annotation (SegUI):
- Platform independent: User interface runs in the browser on any browser capable device (e.g. notebook, tablet, smartphone) and no installation is needed.
- Easy AI Usage: Select segmentation approaches from AI model zoo and apply to image data no matter whether you work with your notebook, tablet or smartphone.
- Import/Export: Share segmentation data with OMERO simplifying collaboration and scientific discussion.
- Smart data model: Undo/redo segmentation changes to flexibly try all features without fearing mistakes.
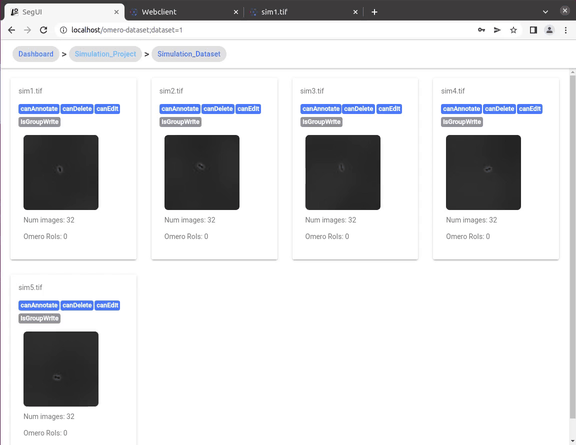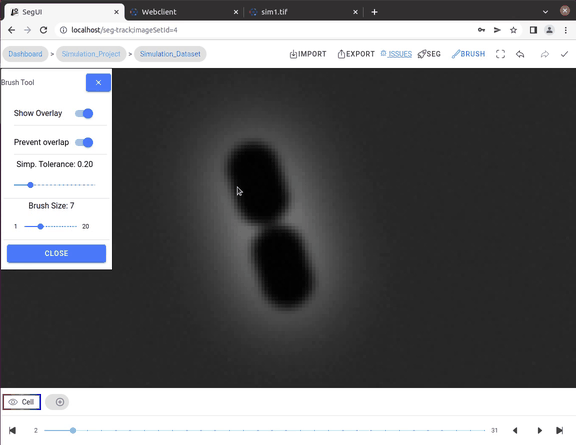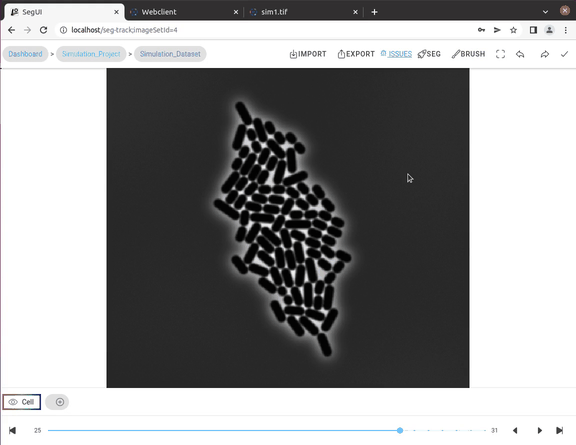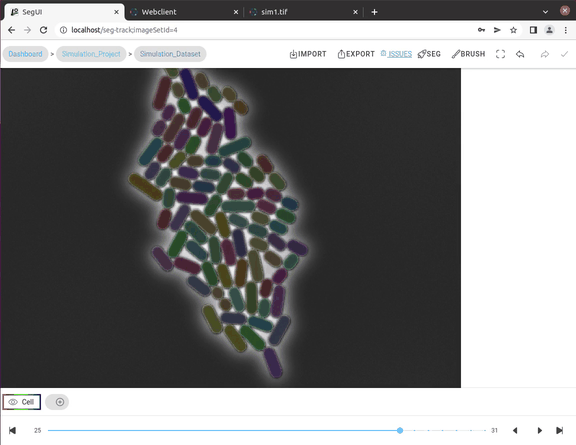
- Manual correction tools: Providing brush tool for drawing segmentations and a multi-select tool for selecting and deleting multiple segmentation contours.
- Multi-label handling: Handles complex segmentation scenarios with multiple annotation labels and also supports AI approaches with multiple annotation classes.
- Automated Saving: Do not loose any progress. We automatically save your work in the background.
- OMERO Integration: User and data management using OMERO.
- Scalability: The docker setup can be scaled from single-users to teams or a full cloud setup - depending on your demand.
- Simple docker installation: Dockers containerization functionalities allow to easily deploy ObiWan Microbi.
- Remote accessibility: You are no longer bound to one workstation. All your data is stored in OMERO and available in your browser after your login.
- Facilitate collaboration: OMERO allows to share data in groups such that ground truth creation can be performed in a team of experts.
This project is licensed under the MIT License - see the LICENSE.md file for details.