Interactive multiscale visualization for structural variation in human genomes.
- Project/Documentation: https://chromoscope.bio
- Demo: https://chromoscope.bio/app/
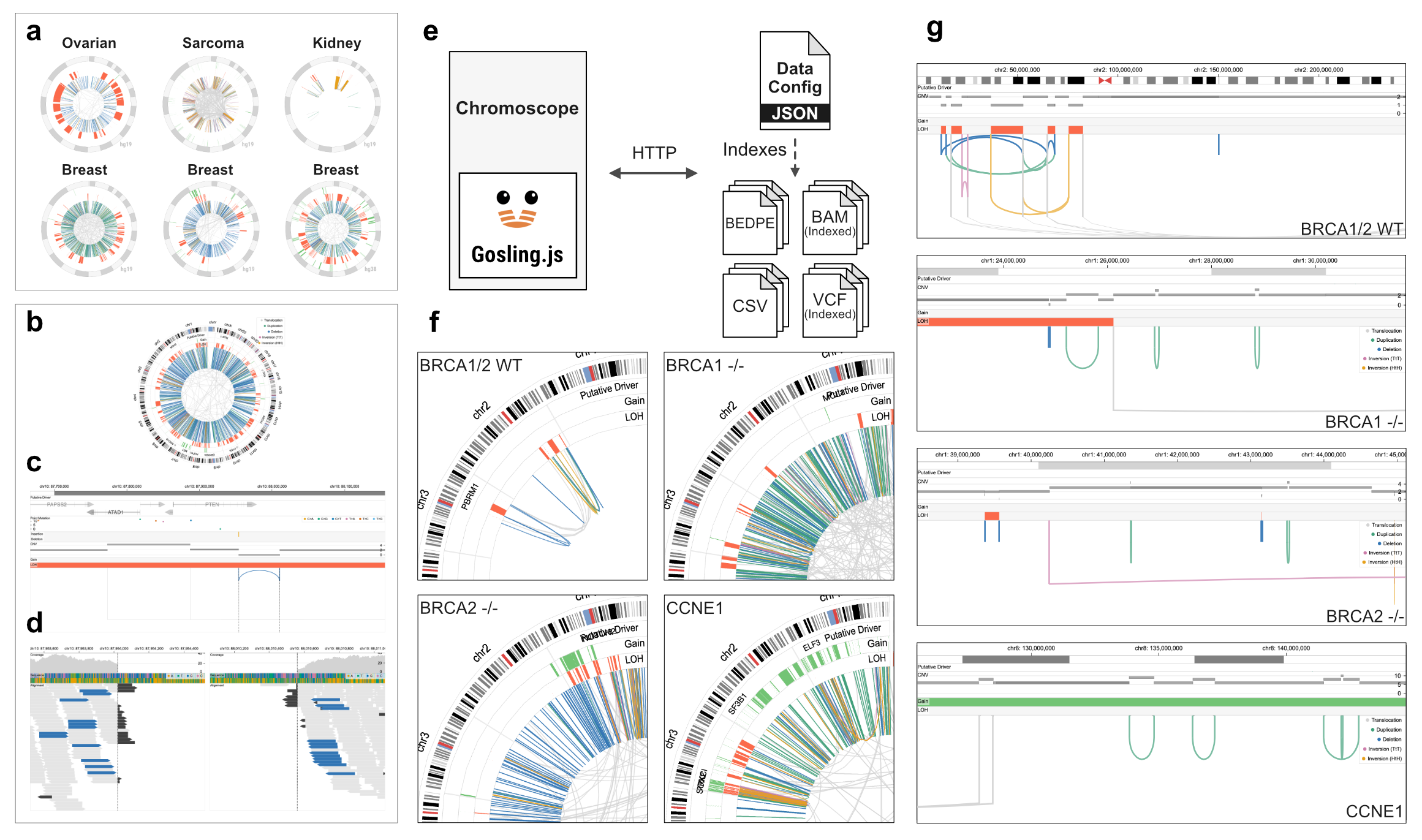
The Chromoscope interface consists of four views for analyzing structural variation in cancer genomes at multiple scales (a–d). Chromoscope uses a "data config" to load datasets via HTTP requests and does not require setting up a server (e). The data can be either stored privately or publicly (e.g., cloud buckets or local servers). Chromoscope captures distinct patterns of structural variations and their copy number footprint in samples with different types of chromosomal instability, such as chromothripsis, or associated with loss of BRCA1 -/-, BRCA2 -/-, or CCNE1 amplification (f-g).
Ask for help and propose or discuss new features. Please email dominik_glodzik@hms.harvard.edu if you would like to join.
Clone this repo on your local machine first.
Move to the project folder and then install all dependencies using Yarn. You need to install Yarn first if you do not have it installed.
cd chromoscope
yarn installRun the local instance.
yarn startOpen the browser to see the local Chromoscope.
http://localhost:3000Install dependencies for the documentation website.
yarn --cwd docs installStart the local server for the documentation.
yarn start-docsOpen the browser to see the local documentation.
http://localhost:3000/docsPlease cite the following publication:
Sehi L’Yi, Dominika Maziec, Victoria Stevens, Trevor Manz, Alexander Veit, Michele Berselli, Peter J. Park, Dominik Głodzik, and Nils Gehlenborg. Chromoscope: interactive multiscale visualization for structural variation in human genomes. Nat Methods 20, 1834–1835 (2023). https://doi.org/10.1038/s41592-023-02056-x
Chromoscope is funded in part through a grant awarded by Innovation in Cancer Informatics.